Background to Shell (no, not ? shells)
An operating system (OS) can be thought of two components, the kernel and shell. The kernel is the core of the OS and handles low-level operations such as starting/ending processes, and allocating memory when you open and close your browser. Jointly, the shell is a program that can send very direct, specific commands to system tasks.
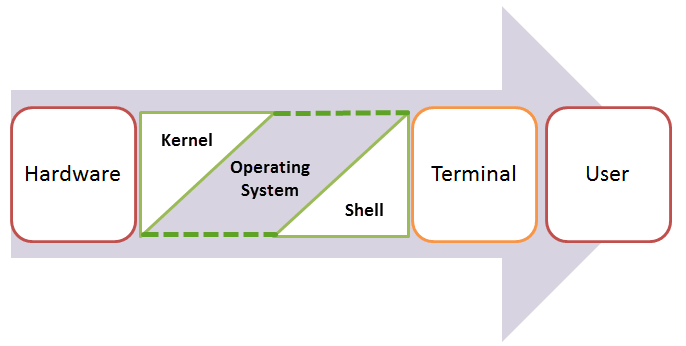
For example, listing all your files in a directory by their date modified. You could have a Graphical User Interface (GUI) program that could do this (finder, explorer) but some commands are so specific that is up to you for figure out how to create the specific command! BASH (Bourne Again Shell) is a shell that is most commonly used in bioinformatics and in programming in general. Mac Users already have BASH installed (open ‘Terminal’; although this may be Zsh soon). Windows users can open up install Ubuntu which has BASH. Alternatively you can login into a Linux server like the BAR or SciNet using PuTTY. Just note that when you login to a remote computer you are accessing that computer’s files!
Essential Commands
Navigating your place
ssh -p 22 neo@thematrix # login as neo to the remote matrix server under port 22 (usually the ssh port), use password prompt ls # print all files/locations less hidden ls -a # print all files including hidden ls -l # print extra info ls -al # combine logic above ls my-directory-somewhere-else/stuff # print stuff in other path pwd # print current directory dir in absolute path mkdir # make a folder in current directory cp doc.txt doc2.txt # make a copy rm file.txt # delete file, PERMANENT mv doc.txt ../. # move this file to the parent directory whoami # print current logged in user
Notice how ls -al I was able to (1) use flags to change the output of the command and (2) combine the flags to give me even more control. In general flags are a gigantic part of running shell commands (and bioinformatics programs) and you should get familiar with flags by checking out each commands man page by using man ls for example.
File permissions
One concept you should understand (among many) is that files may be readable (r), writable (w) and executable (x). And that these specific permissions can be assigned to Owner (usually a sys admin), Group (list of special accounts) and Others (all users) respectively. You can get the file permissions by using ls -l. You can also change the permissions using chmod #chmod 777 myFile.txt. Cleverly, the permissions to the above groups can be summarized in a binary number (see below).

That being said, we should have made the BAR fairly secure in that you cannot delete others’ or sensitive files but always make sure you are know what you are doing because once you delete something in bash (using rm), IT CANNOT BE UNDONE!
String processing (Intro)
One of the largest advantages of using BASH is its powerful arsenal of string manipulation tools such as being able to quickly split fields (columns) of a comma separated values (CSV) file into a more workable format and using regular expressions to parse and replace text efficiently. I highlight this for those undergraduates who will be doing a lot of text/data parsing (working with DNA sequences, large tables [CSVs] for example). For those students, look into tools such as:
head file.txt #print top lines tail file.txt #print last lines vim newFile.txt #use the vim text editor to edit your file nano newFile.txt #use nano text editor grep 'word' file.txt #find 'word' in your file and print the lines, NB many options here! sort file.txt #sort this file, according to your criteria diff file1. txt file2.txt #print the different lines in this file, very useful for code merging (like git merge)
Shell Scripting
You can write shell scripts (files ending in .sh) that are quite useful for automating tasks. Moreover for those using a cluster you will need to write simple shell scripts that will need to be executed. Here is an example shell script but know that you can also run these commands line-by-line in your console. The first line is a shebang tells what shell to execute the script with. Note that the syntax for shell scripts is very strict so be patient.
#!/bin/bash echo "Starting script!" # log to console a=5 b=10 echo $(( $a + $b )) python myPyScript.py #run python script
Useful tips
- Combine output of one command to another using ‘|’ (pipe). The below code takes the output of
lsand prints the top 5 lines/files. A usecase for this may be if you have a large directory and don’t want a large output.ls -l | head -5
- Write the output of a shell command (or multiple like above) to a file using ‘>’. Below makes a text file with current date.
date > example.txt
- For those running code that may take a while (and IS NOT using a job submitter on a cluster like SciNet), you can run a screen and allow that process to execute. When it is finished you can return back to the screen. Alternative it is quite nice for going back to where you left off earlier. I use this to run simple webservers sometimes, I go to that screen when I need to debug HTTP requests.
screen -S running_my_long_python_jobThis will start a screen which you can execute your python script, then detach from and logout from your server viaCtrl+adandexitrespectively.
- Set alias for commands you use a lot.
alias lsal="ls -al"This will set an alias such that when you executelsalit will execute the right hand assigned command. Note that this will get erased after you exit your session. To set it permanently you should modify your .bashrc or .bash_profile which gets run everytime you re-enter your shell.